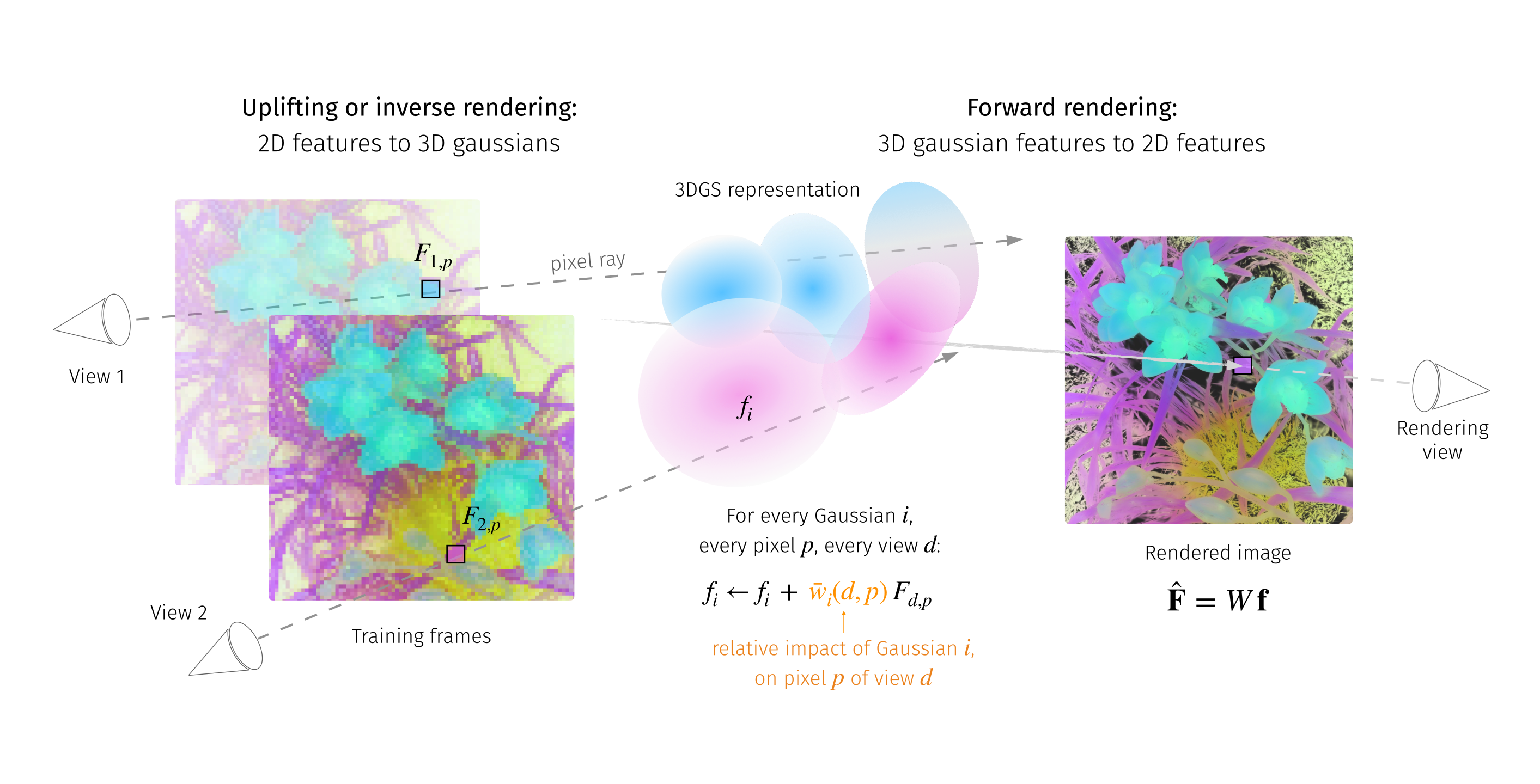
LUDVIG: Learning-free Uplifting of 2D Visual features to Gaussian Splatting scenes
Recently, I worked on transferring representations from DINOv2, SAM, and CLIP into 3D Gaussian Splatting scenes. My study showed that a simple aggregation of 2D features is highly effective, achieving competitive results on segmentation and detection tasks while providing significant speed-ups over prior methods minimizing a reprojection loss. For 3D segmentation, we introduce a graph diffusion mechanism that enriches 3D features, such as coarse segmentation masks, by leveraging 3D geometry and pairwise similarities induced by DINOv2.
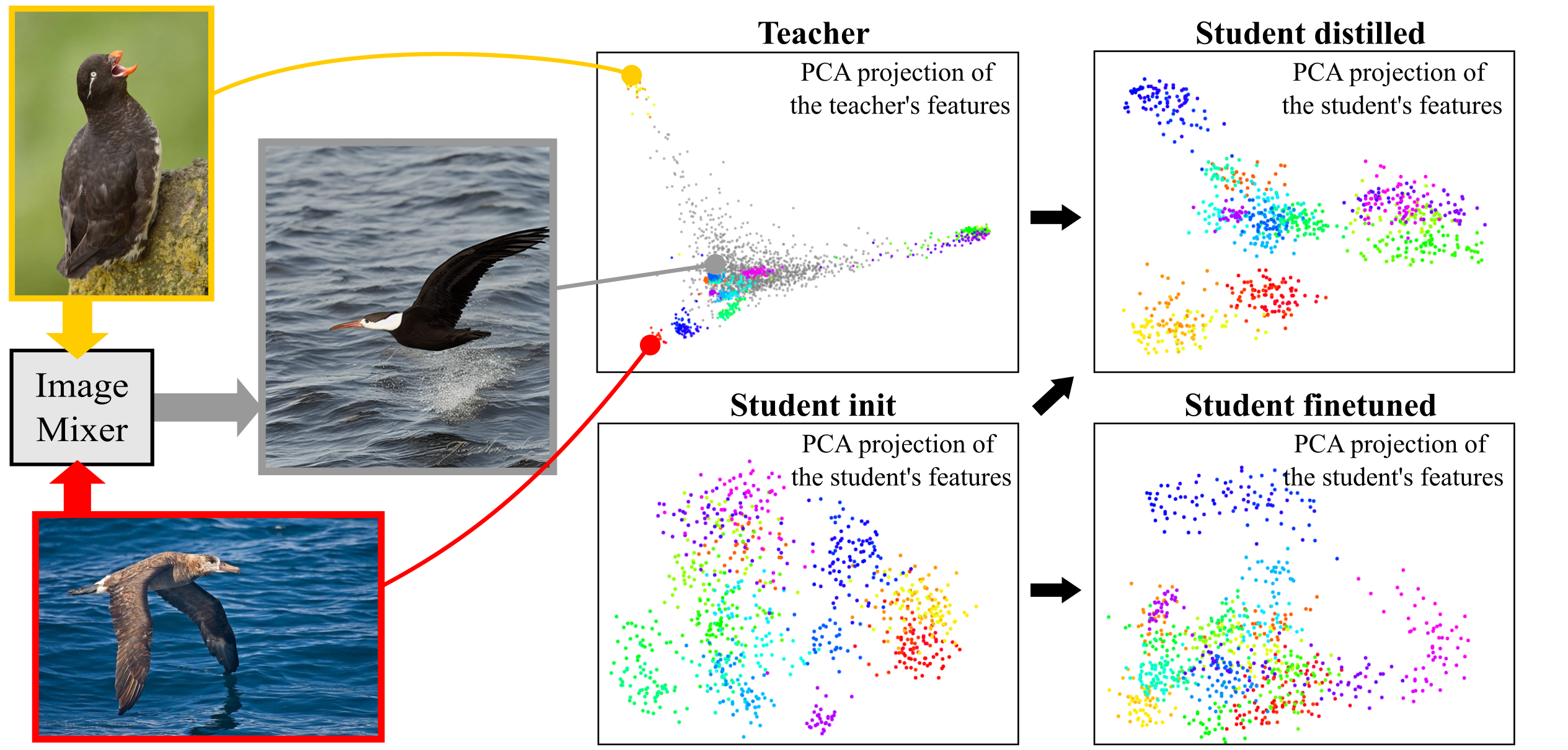
On Good Practices for Task-Specific Distillation of Large Pretrained Visual Models
My second PhD project (TMLR 2024) delineates good practices for leveraging large pretrained visual models to train smaller models on specific tasks.
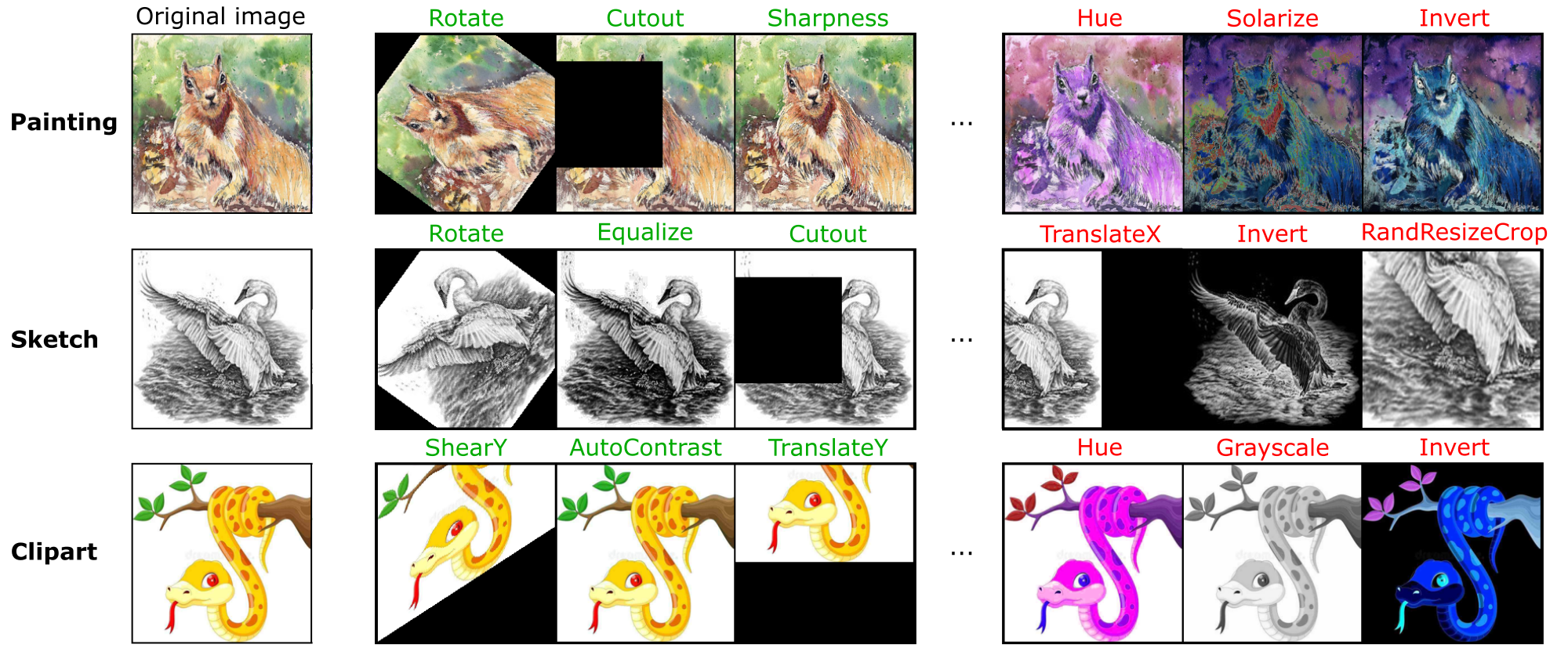
SLACK: Stable Learning of Augmentations with Cold-start and KL regularization
My first PhD project (CVPR 2023) focused on automatically learning optimal data augmentation policies using bilevel optimization.
Self-supervised learning from 3D medical images (Philips Research, Paris)
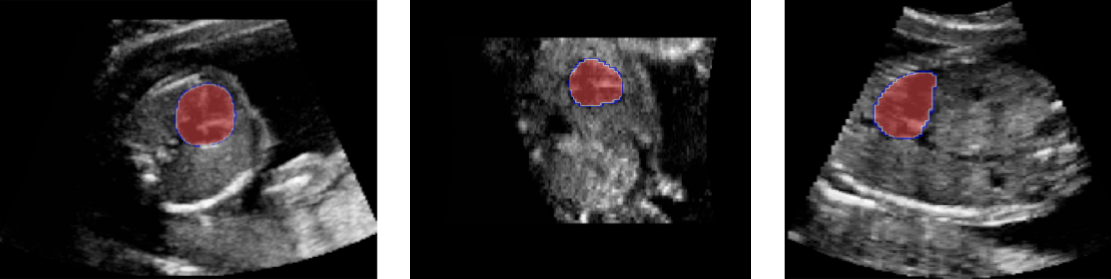
In 2021, I completed a 6-month internship at Philips Research (France), where I worked on self-supervised learning from 3D medical images, with evaluation on 3D ultrasound image segmentation and 3D CT scan classification.
Phylogenetic tree reconstruction (Weill Cornell, New York)
In 2020, I completed a 6-month internship in the Landau Lab (Weill Cornell Medicine and the NYGC, New York), where I worked on reconstructing phylogenetic trees (of cell divisions) based on mutations observed in microsatellite sequences obtained through single-cell RNA sequencing.

Shape optimization with Geometric Deep Learning (Neural Concept, Lausanne)
In 2019, I completed a 6-month internship at Neural Concept (start-up in EPFL, Lausanne), where I worked on optimizing 3D shapes (e.g. with respect to the lift-to-drag ratio) using a Geometric Deep Learning model trained on the outputs from physical simulations.